Reference-based cell-type annotation can significantly reduce time and effort in single-cell analysis by transferring labels from a previously-annotated dataset to a new dataset. However, label transfer by end-to-end computational methods is challenging due to the entanglement of technical (e.g., from different sequencing batches or techniques) and biological (e.g., from different cellular microenvironments) variation, only the first of which must be removed.
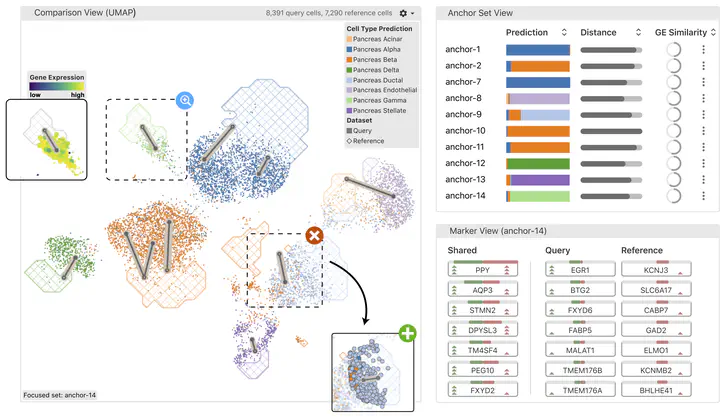
To address this issue, we propose Polyphony, an interactive transfer learning (ITL) framework, to complement biologists’ knowledge with advanced computational methods. Polyphony is motivated and guided by domain experts’ needs for a controllable, interactive, and algorithm-assisted annotation process, identified through our multi-round expert interviews with seven biologists. We introduce anchors, analogous cell populations across datasets, as a paradigm to explain the computational process and collect user feedback for model improvement. A set of visualizations and interactions is provided to empower users to add, delete, or modify anchors, resulting in refined cell type annotations. We demonstrate the effectiveness of this approach through quantitative experiments, two hypothetical use cases, and interviews with two biologists. The results show that our anchor-based ITL method takes advantage of both human and machine intelligence in annotating massive single-cell datasets.